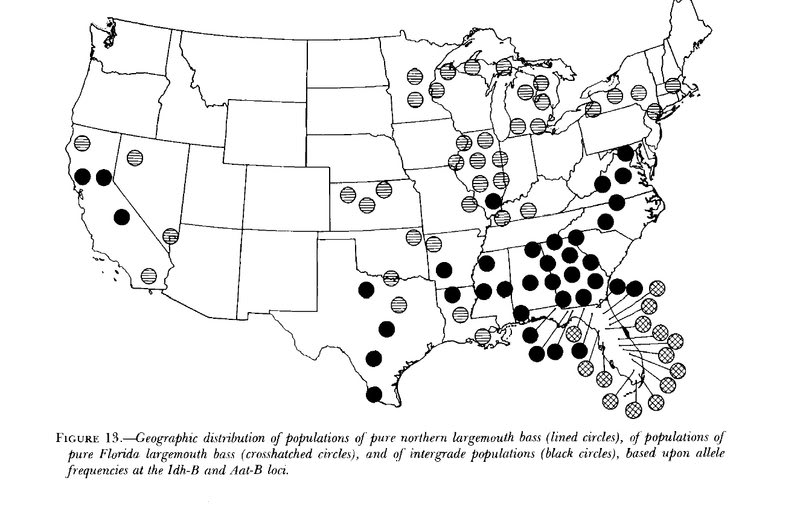
There is a lot unknown about the subject of your question. There are some threads here on many aspects of that goal. Read up on catchability and growth rates. Look into the research on TPWD share a lunker genetics and studies on integration of Fla and Northern LMB in TX waters. Here is a bit to ponder.
Subspecies Composition of Angled and Electrofished Largemouth Bass in Texas Reservoirs , Dijar J. Lutz-Carrillo, and Spencer Dumont in Proceedings of the Annual Conference of the Southeast Association of Fish and Wildlife. Agencies 66:75–81, 2012 Proc . The study’s results provide biologists with a provocative concept that, in southern waters (natural integrated zone of FLMB and NLMB), FLMB likely are more difficult to angle than NLMB, but the phenotype (genetic trait) of reduced angler susceptibility (catchability) is mitigated (reduced) by introgression (genetic mixing), even at low levels of NLMB genes. While many factors may function concurrently to determine angling susceptibility, including fishing pressure, naivety of individual fish, learned lure avoidance, and stress from catch and release , we now know that catchability is a genetic and heritable trait as was previously discussed in earlier Cutting Edge articles ( see Garrett (2002) and Philipp et al. (2009) ). Note that when pure FLMB were removed from the dataset numbers (leaving only crosses with high levels of FLMB genetic influence) the trait of reduced catchability disappeared. Stated differently the addition of only a small amount of NLMB genetics caused poor catchability to disappear.
Trophy fish (≥13 lbs.) from introgressed populations are represented disproportionately as non-introgressed FLMB relative to the frequency of FLMB in the general population ( TPWD, unpublished data). Thus, in an appropriate environment, genetic composition appears to be a critical component of maximum size in largemouth bass. While most of the differences in growth and maximum size between these subspecies are likely due to intrinsic physiological differences, behavioral differences, such as reduced angling susceptibility, may also play a role. FLMB that are potentially less susceptible to angling would be removed from populations at reduced rates relative to non-introgressed NLMB or crosses, allowing a greater proportion of FLMB to reach an older age and greater size. However I recall TPWD data also showing many of the share-a-lunker fish were high FLMB percentage crosses. From Pond Boss – The Cutting Edge.
From 2010-2012, fifteen trophy-sized hatchery coded wire tag (CWT) bass weighing 8-12.5 lbs were recaptured during FWC long term monitoring samples or largemouth bass angler tournaments. Furthermore, 77 largemouth bass weighing 8-15 lbs were collected in 2013, of which only two were not identified as pure Floridanus based on genetic micro-satellite analysis of fin clips. Most of these are likely hatchery reared fish as prior to any stocking, the percentage of pure Floridanus in Lake Talquin was reported at 12%. These long-term data suggest that the stocking of advanced fingerling Florida largemouth bass reared on live prey into a recruitment limited system with abundant, appropriate size forage improved trophy largemouth bass abundance and enhanced the quality of a trophy bass fishery.
The field of conservation genetics, harnessing the power of next-generation sequencing, is rapidly adapting SNP technology (see below note * ) due the advantages of SNP markers including abundance, even genome distribution, ease of multiplexing and lab-to-lab reproducibility. Here we sequenced the transcriptomes of M. salmoides, M. floridanus and their F1 hybrid and identified a set of 3,674 SNP markers with putative fixed allelic differences between species from 2,112 unique genes. We have developed two multiplex panels of 62 SNP markers for the MassARRAY platform and validated their capacity for assessing integrity and hybridization in hatchery and wild populations of bass (n=700). Our data indicates a complexity in natural intergrade populations overlooked with previous marker systems.
• Note
Single Nucleotide Polymorphism (SNP), a variation at a single site in DNA, is the most frequent type of variation in the genome.
The basic principles of SNP array are the same as the DNA microarray. These are the convergence of DNA hybridization, fluorescence microscopy, and solid surface DNA capture. The three mandatory components of the SNP arrays are:
1. An array containing immobilized allele-specific oligonucleotide (ASO) probes.
2. Fragmented nucleic acid sequences of target, labeled with fluorescent dyes.
3. A detection system that records and interprets the hybridization signal.
The ASO probes are often chosen based on sequencing of a representative panel of individuals: positions found to vary in the panel at a specified frequency are used as the basis for probes. SNP chips are generally described by the number of SNP positions they assay. Two probes must be used for each SNP position to detect both alleles; if only one probe were used, experimental failure would be indistinguishable from homozygosity of the non-probed allele.